Introduction
Linear regression and ridge regression are simple machine learning techniques that aim to estimate the parameters of a linear model. Assuming we have 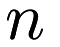 predictor points
predictor points 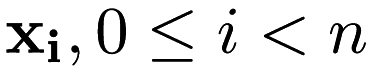 of dimensionality
of dimensionality 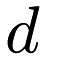 and
and 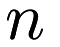 responses
responses 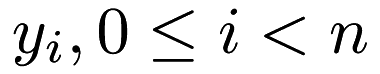 , we are trying to estimate the best fit for
, we are trying to estimate the best fit for 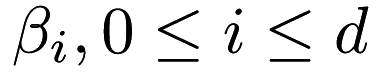 in the linear model
in the linear model
![\[ y_i = \beta_0 + \displaystyle\sum_{j = 1}^{d} \beta_j x_{ij} \]](form_216.png)
for each predictor 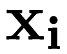 and response
and response 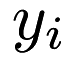 . If we take each predictor
. If we take each predictor 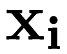 as a row in the matrix
as a row in the matrix 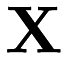 and each response
and each response 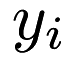 as an entry of the vector
as an entry of the vector 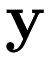 , we can represent the model in vector form:
, we can represent the model in vector form:
![\[ \mathbf{y} = \mathbf{X} \mathbf{\beta} + \beta_0 \]](form_221.png)
The result of this method is the vector  , including the offset term (or intercept term)
, including the offset term (or intercept term) 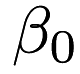 .
.
mlpack provides:
- a simple command-line executable to perform linear regression or ridge regression
- a simple C++ interface to perform linear regression or ridge regression
Table of Contents
A list of all the sections this tutorial contains.
- Introduction
- Table of Contents
- Command-Line 'mlpack_linear_regression'
- The 'LinearRegression' class
- Further documentation
Command-Line 'mlpack_linear_regression'
The simplest way to perform linear regression or ridge regression in mlpack is to use the mlpack_linear_regression executable. This program will perform linear regression and place the resultant coefficients into one file.
The output file holds a vector of coefficients in increasing order of dimension; that is, the offset term ( 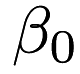 ), the coefficient for dimension 1 (
), the coefficient for dimension 1 ( 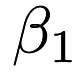 , then dimension 2 (
, then dimension 2 ( 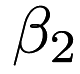 ) and so forth, as well as the intercept. This executable can also predict the
) and so forth, as well as the intercept. This executable can also predict the 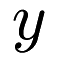 values of a second dataset based on the computed coefficients.
values of a second dataset based on the computed coefficients.
Below are several examples of simple usage (and the resultant output). The option is used so that verbose output is given. Further documentation on each individual option can be found by typing
One file, generating the function coefficients
Convenient program timers are given for different parts of the calculation at the bottom of the output, as well as the parameters the simulation was run with. Now, if we look at the output model file, which is lr.xml,
As you can see, the function for this input is 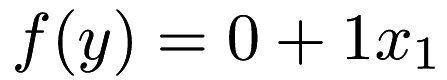 . We can see that the model we have trained catches this; in the
. We can see that the model we have trained catches this; in the <parameters> section of lr.xml, we can see that there are two elements, which are (approximately) 0 and 1. The first element corresponds to the intercept 0, and the second column corresponds to the coefficient 1 for the variable 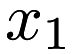 . Note that in this example, the regressors for the dataset are the second column. That is, the dataset is one dimensional, and the last column has the
. Note that in this example, the regressors for the dataset are the second column. That is, the dataset is one dimensional, and the last column has the 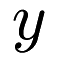 values, or responses, for each row. You can specify these responses in a separate file if you want, using the
values, or responses, for each row. You can specify these responses in a separate file if you want, using the –input_responses, or -r, option.
Train a multivariate linear regression model
Multivariate linear regression means that the response variable is predicted by more than just one input variable. In this example we will try to fit a multivariate linear regression model to data that contains four variables, stored in dataset_2.csv.
Now let's run mlpack_linear_regression as usual:
If we take a look at the lr.xml output we can see the <parameters> part has five elements which the first corresponds to 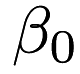 , the second corresponds to
, the second corresponds to 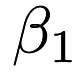 , and so on. This is equivalent to
, and so on. This is equivalent to 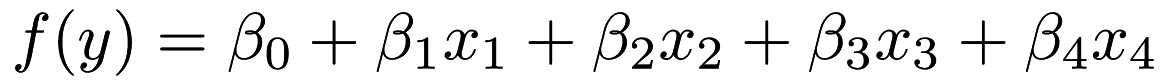 or
or 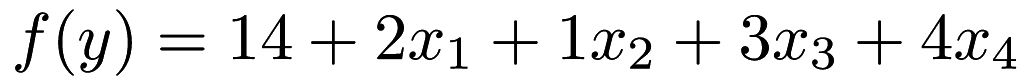 .
.
Compute model and predict at the same time
We used the same dataset, so we got the same parameters. The key thing to note about the predict.csv dataset is that it has the same dimensionality as the dataset used to create the model, one. If the model generating dataset has 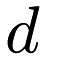 dimensions, so must the dataset we want to predict for.
dimensions, so must the dataset we want to predict for.
Prediction using a precomputed model
Using ridge regression
Sometimes, the input matrix of predictors has a covariance matrix that is not invertible, or the system is overdetermined. In this case, ridge regression is useful: it adds a normalization term to the covariance matrix to make it invertible. Ridge regression is a standard technique and documentation for the mathematics behind it can be found anywhere on the Internet. In short, the covariance matrix
![\[ \mathbf{X}' \mathbf{X} \]](form_231.png)
is replaced with
![\[ \mathbf{X}' \mathbf{X} + \lambda \mathbf{I} \]](form_232.png)
where 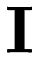 is the identity matrix. So, a
is the identity matrix. So, a 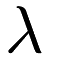 parameter greater than zero should be specified to perform ridge regression, using the
parameter greater than zero should be specified to perform ridge regression, using the –lambda (or -l) option. An example is given below.
Further documentation on options should be found by using the –help option.
The 'LinearRegression' class
The 'LinearRegression' class is a simple implementation of linear regression.
Using the LinearRegression class is very simple. It has two available constructors; one for generating a model from a matrix of predictors and a vector of responses, and one for loading an already computed model from a given file.
The class provides one method that performs computation:
Once you have generated or loaded a model, you can call this method and pass it a matrix of data points to predict values for using the model. The second parameter, predictions, will be modified to contain the predicted values corresponding to each row of the points matrix.
Generating a model
Setting a model
Assuming you already have a model and do not need to create one, this is how you would set the parameters for a LinearRegression instance.
Load a model from a file
If you have a generated model in a file somewhere you would like to load and use, you can use data::Load() to load it.
Prediction
Once you have generated or loaded a model using one of the aforementioned methods, you can predict values for a dataset.
Setting lambda for ridge regression
As discussed in Prediction using a precomputed model, ridge regression is useful when the covariance of the predictors is not invertible. The standard constructor can be used to set a value of lambda:
In addition, the Lambda() function can be used to get or modify the lambda value:
Further documentation
For further documentation on the LinearRegression class, consult the complete API documentation.


