Introduction
DETs perform the unsupervised task of density estimation using decision trees. Using a trained density estimation tree (DET), the density at any particular point can be estimated very quickly (O(log n) time, where n is the number of points the tree is built on).
The details of this work is presented in the following paper:
mlpack provides:
- a simple command-line executable to perform density estimation and related analyses using DETs
- a generic C++ class (DTree) which provides various functionality for the DETs
- a set of functions in the namespace mlpack::det to perform cross-validation for the task of density estimation with DETs
Table of Contents
A list of all the sections this tutorial contains.
- Introduction
- Table of Contents
- Command-Line mlpack_det
- The 'DTree' class
- 'namespace mlpack::det'
- Further Documentation
Command-Line mlpack_det
The command line arguments of this program can be viewed using the -h option:
Plain-vanilla density estimation
We can just train a DET on the provided data set S. Like all datasets mlpack uses, the data should be row-major (mlpack transposes data when it is loaded; internally, the data is column-major – see this page for more information).
By default, mlpack_det performs 10-fold cross-validation (using the 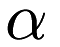 -pruning regularization for decision trees). To perform LOOCV (leave-one-out cross-validation), which can provide better results but will take longer, use the following command:
-pruning regularization for decision trees). To perform LOOCV (leave-one-out cross-validation), which can provide better results but will take longer, use the following command:
To perform k-fold crossvalidation, use -f k (or –folds k). There are certain other options available for training. For example, in the construction of the initial tree, you can specify the maximum and minimum leaf sizes. By default, they are 10 and 5 respectively; you can set them using the -M (–max_leaf_size) and the -N (–min_leaf_size) options.
In case you want to output the density estimates at the points in the training set, use the -e (–training_set_estimates_file) option to specify the output file to which the estimates will be saved. The first line in density_estimates.txt will correspond to the density at the first point in the training set. Note that the logarithm of the density estimates are given, which allows smaller estimates to be saved.
Estimation on a test set
Often, it is useful to train a density estimation tree on a training set and then obtain density estimates from the learned estimator for a separate set of test points. The -T (–test_file) option allows specification of a set of test points, and the -E (–test_set_estimates_file) option allows specification of the file into which the test set estimates are saved. Note that the logarithm of the density estimates are saved; this allows smaller values to be saved.
Computing the variable importance
The variable importance (with respect to density estimation) of the different features in the data set can be obtained by using the -i (–vi_file ) option. This outputs the absolute (as opposed to relative) variable importance of the all the features into the specified file.
Saving trained DETs
The mlpack_det program is capable of saving a trained DET to a file for later usage. The –output_model_file or -M option allows specification of the file to save to. In the example below, a DET trained on dataset.csv is saved to the file det.xml.
Loading trained DETs
A saved DET can be used to perform any of the functionality in the examples above. A saved DET is loaded with the –input_model_file or -m option. The example below loads a saved DET from det.xml and outputs density estimates on the dataset test_dataset.csv into the file estimates.csv.
The 'DTree' class
This class implements density estimation trees. Below is a simple example which initializes a density estimation tree.
Public Functions
The function Grow() greedily grows the tree, adding new points to the tree. Note that the points in the dataset will be reordered. This should only be run on a tree which has not already been built. In general, it is more useful to use the Trainer() function found in 'namespace mlpack::det'.
Note that the alternate volume regularization should not be used (see ticket #238).
To estimate the density at a given query point, use the following code. Note that the logarithm of the density is returned.
Computing the variable importance of each feature for the given DET.
'namespace mlpack::det'
The functions in this namespace allows the user to perform tasks with the 'DTree' class. Most importantly, the Trainer() method allows the full training of a density estimation tree with cross-validation. There are also utility functions which allow printing of leaf membership and variable importance.
Utility Functions
The code below details how to train a density estimation tree with cross-validation.
Note that the alternate volume regularization should be set to false because it has known bugs (see #238).
To print the class membership of leaves in the tree into a file, see the following code.
Note that you can find the number of classes with max(labels) + 1. The variable importance can also be printed to a file in a similar manner.
Further Documentation
For further documentation on the DTree class, consult the complete API documentation.


